Medicare data
<data.medicare.gov/Hospital-Compare>: A web site suggested by Kim Druschel at Saint Louis University
One dataset there is about infections associated with hospitalization. https://data.medicare.gov/Hospital-Compare/Healthcare-Associated-Infections-Hospital/77hc-ibv8
- Download as CSV
- In RStudio, go to Environment/ImportDataset
Wrangling data:
library(dplyr, quietly = TRUE, verbose = FALSE)
library(readr, quietly = TRUE, verbose = FALSE)
Infections <- readr::read_csv("~/Downloads/Healthcare_Associated_Infections_-_Hospital.csv")## Parsed with column specification:
## cols(
## `Provider ID` = col_character(),
## `Hospital Name` = col_character(),
## Address = col_character(),
## City = col_character(),
## State = col_character(),
## `ZIP Code` = col_double(),
## `County Name` = col_character(),
## `Phone Number` = col_double(),
## `Measure Name` = col_character(),
## `Measure ID` = col_character(),
## `Compared to National` = col_character(),
## Score = col_character(),
## Footnote = col_character(),
## `Measure Start Date` = col_character(),
## `Measure End Date` = col_character(),
## Location = col_character()
## )unique(Infections$`Measure Name`)## [1] "MRSA Bacteremia"
## [2] "Clostridium Difficile (C.Diff): Patient Days"
## [3] "Central Line Associated Bloodstream Infection: Number of Device Days"
## [4] "SSI - Abdominal Hysterectomy"
## [5] "Catheter Associated Urinary Tract Infections (ICU + select Wards): Predicted Cases"
## [6] "Central Line Associated Bloodstream Infection (ICU + select Wards): Lower Confidence Limit"
## [7] "Catheter Associated Urinary Tract Infections (ICU + select Wards): Lower Confidence Limit"
## [8] "SSI - Colon Surgery"
## [9] "SSI - Colon Surgery: Upper Confidence Limit"
## [10] "Central Line Associated Bloodstream Infection (ICU + select Wards): Predicted Cases"
## [11] "Catheter Associated Urinary Tract Infections (ICU + select Wards)"
## [12] "MRSA Bacteremia: Predicted Cases"
## [13] "SSI - Abdominal Hysterectomy: Upper Confidence Limit"
## [14] "Clostridium Difficile (C.Diff): Upper Confidence Limit"
## [15] "MRSA Bacteremia: Observed Cases"
## [16] "MRSA Bacteremia: Patient Days"
## [17] "SSI - Abdominal Hysterectomy: Predicted Cases"
## [18] "Central Line Associated Bloodstream Infection (ICU + select Wards): Observed Cases"
## [19] "SSI - Abdominal Hysterectomy: Lower Confidence Limit"
## [20] "Clostridium Difficile (C.Diff)"
## [21] "Central Line Associated Bloodstream Infection (ICU + select Wards): Upper Confidence Limit"
## [22] "Central Line Associated Bloodstream Infection (ICU + select Wards)"
## [23] "Catheter Associated Urinary Tract Infections (ICU + select Wards): Upper Confidence Limit"
## [24] "Catheter Associated Urinary Tract Infections (ICU + select Wards): Number of Urinary Catheter Days"
## [25] "Catheter Associated Urinary Tract Infections (ICU + select Wards): Observed Cases"
## [26] "SSI - Colon Surgery: Lower Confidence Limit"
## [27] "SSI - Colon Surgery: Number of Procedures"
## [28] "SSI - Colon Surgery: Predicted Cases"
## [29] "SSI - Colon Surgery: Observed Cases"
## [30] "SSI - Abdominal Hysterectomy: Number of Procedures"
## [31] "SSI - Abdominal Hysterectomy: Observed Cases"
## [32] "MRSA Bacteremia: Lower Confidence Limit"
## [33] "MRSA Bacteremia: Upper Confidence Limit"
## [34] "Clostridium Difficile (C.Diff): Lower Confidence Limit"
## [35] "Clostridium Difficile (C.Diff): Predicted Cases"
## [36] "Clostridium Difficile (C.Diff): Observed Cases"# separate institution data from measurements
measurements <- names(Infections)[9:15]
Institutions <-
Infections %>%
select( - one_of(measurements)) %>%
group_by(`Provider ID`) %>%
filter(row_number() == 1)
Measures <-
Infections %>%
select(`Provider ID`, one_of(measurements))Let’s look at central line associated bloodstream infections.
Central_Line <-
Measures %>%
filter(grepl("Central Line Associated", `Measure Name`)) %>%
mutate(Score = ifelse(Score %in% c("--", "Not Available"), NA, parse_number(Score))) %>%
mutate(condition = gsub("HAI_._", "", `Measure ID`)) %>%
select(-`Measure Start Date`, -`Measure End Date`, -`Footnote`,
-`Compared to National`, -`Measure Name`, -`Measure ID`) %>%
tidyr::spread(key = condition, value = Score)## Warning: 11908 parsing failures.
## row col expected actual
## 1 -- a number Not Available
## 2 -- a number Not Available
## 3 -- a number Not Available
## 4 -- a number Not Available
## 5 -- a number Not Available
## ... ... ........ .............
## See problems(...) for more details.Abdominal <-
Measures %>%
filter(grepl("SSI - Abdom", `Measure Name`)) %>%
mutate(Score = ifelse(Score %in% c("--", "Not Available"), NA, parse_number(Score))) %>%
mutate(condition = gsub("HAI_._", "", `Measure ID`)) %>%
select(-`Measure Start Date`, -`Measure End Date`, -`Footnote`,
-`Compared to National`, -`Measure Name`, -`Measure ID`) %>%
tidyr::spread(key = condition, value = Score)## Warning: 17871 parsing failures.
## row col expected actual
## 1 -- a number Not Available
## 2 -- a number Not Available
## 3 -- a number Not Available
## 4 -- a number Not Available
## 5 -- a number Not Available
## ... ... ........ .............
## See problems(...) for more details.Colon <-
Measures %>%
filter(grepl("SSI - Colon", `Measure Name`)) %>%
mutate(Score = ifelse(Score %in% c("--", "Not Available"), NA, parse_number(Score))) %>%
mutate(condition = gsub("HAI_._", "", `Measure ID`)) %>%
select(-`Measure Start Date`, -`Measure End Date`, -`Footnote`,
-`Compared to National`, -`Measure Name`, -`Measure ID`) %>%
tidyr::spread(key = condition, value = Score)## Warning: 13957 parsing failures.
## row col expected actual
## 1 -- a number Not Available
## 2 -- a number Not Available
## 3 -- a number Not Available
## 4 -- a number Not Available
## 11 -- a number Not Available
## ... ... ........ .............
## See problems(...) for more details.MRSA <-
Measures %>%
filter(grepl("MRSA", `Measure Name`)) %>%
mutate(Score = ifelse(Score %in% c("--", "Not Available"), NA, parse_number(Score))) %>%
mutate(condition = gsub("HAI_._", "", `Measure ID`)) %>%
select(-`Measure Start Date`, -`Measure End Date`, -`Footnote`,
-`Compared to National`, -`Measure Name`, -`Measure ID`) %>%
tidyr::spread(key = condition, value = Score)## Warning: 12046 parsing failures.
## row col expected actual
## 1 -- a number Not Available
## 2 -- a number Not Available
## 4 -- a number Not Available
## 7 -- a number Not Available
## 14 -- a number Not Available
## ... ... ........ .............
## See problems(...) for more details.Catheter <-
Measures %>%
filter(grepl("Catheter", `Measure Name`)) %>%
mutate(Score = ifelse(Score %in% c("--", "Not Available"), NA, parse_number(Score))) %>%
mutate(condition = gsub("HAI_._", "", `Measure ID`)) %>%
select(-`Measure Start Date`, -`Measure End Date`, -`Footnote`,
-`Compared to National`, -`Measure Name`, -`Measure ID`) %>%
tidyr::spread(key = condition, value = Score)## Warning: 10445 parsing failures.
## row col expected actual
## 2 -- a number Not Available
## 3 -- a number Not Available
## 4 -- a number Not Available
## 6 -- a number Not Available
## 7 -- a number Not Available
## ... ... ........ .............
## See problems(...) for more details.C_DIFF <-
Measures %>%
filter(grepl("Clostridium", `Measure Name`)) %>%
mutate(Score = ifelse(Score %in% c("--", "Not Available"), NA, parse_number(Score))) %>%
mutate(condition = gsub("HAI_._", "", `Measure ID`)) %>%
select(-`Measure Start Date`, -`Measure End Date`, -`Footnote`,
-`Compared to National`, -`Measure Name`, -`Measure ID`) %>%
tidyr::spread(key = condition, value = Score)## Warning: 7595 parsing failures.
## row col expected actual
## 2 -- a number Not Available
## 3 -- a number Not Available
## 10 -- a number Not Available
## 11 -- a number Not Available
## 12 -- a number Not Available
## ... ... ........ .............
## See problems(...) for more details.Can we make sense of the variables? Hypotheses …
ELIGCASESrefers to a model output based on the number of patient daysDOPC
library(ggformula, quietly = TRUE, verbose = FALSE)
gf_point(ELIGCASES ~ DOPC, data = Central_Line)## Warning: Removed 1089 rows containing missing values (geom_point).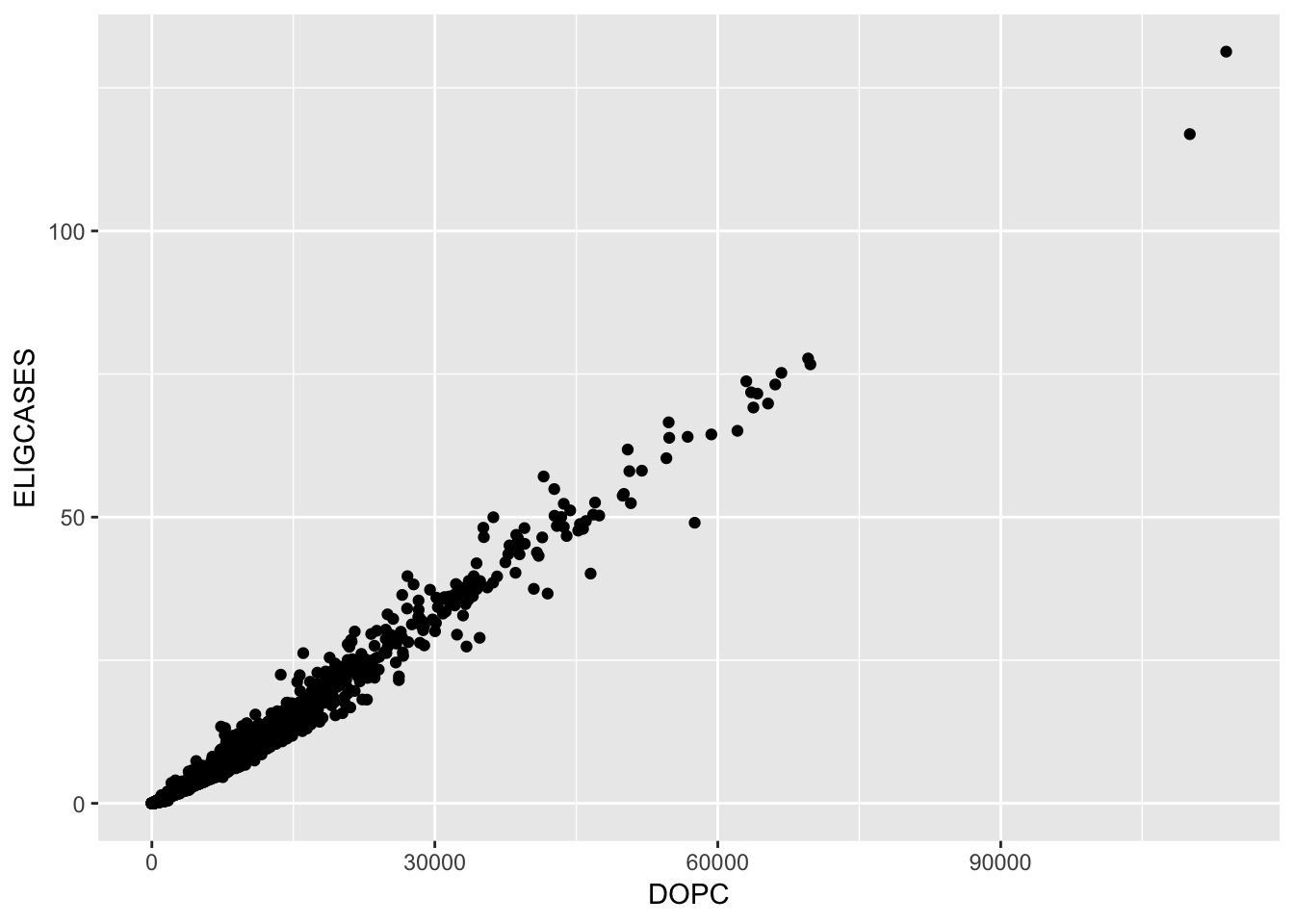
gf_point(NUMERATOR ~ ELIGCASES, data = Central_Line) %>%
gf_abline(intercept = 0, slope = 1, color = "red")## Warning: Removed 1089 rows containing missing values (geom_point).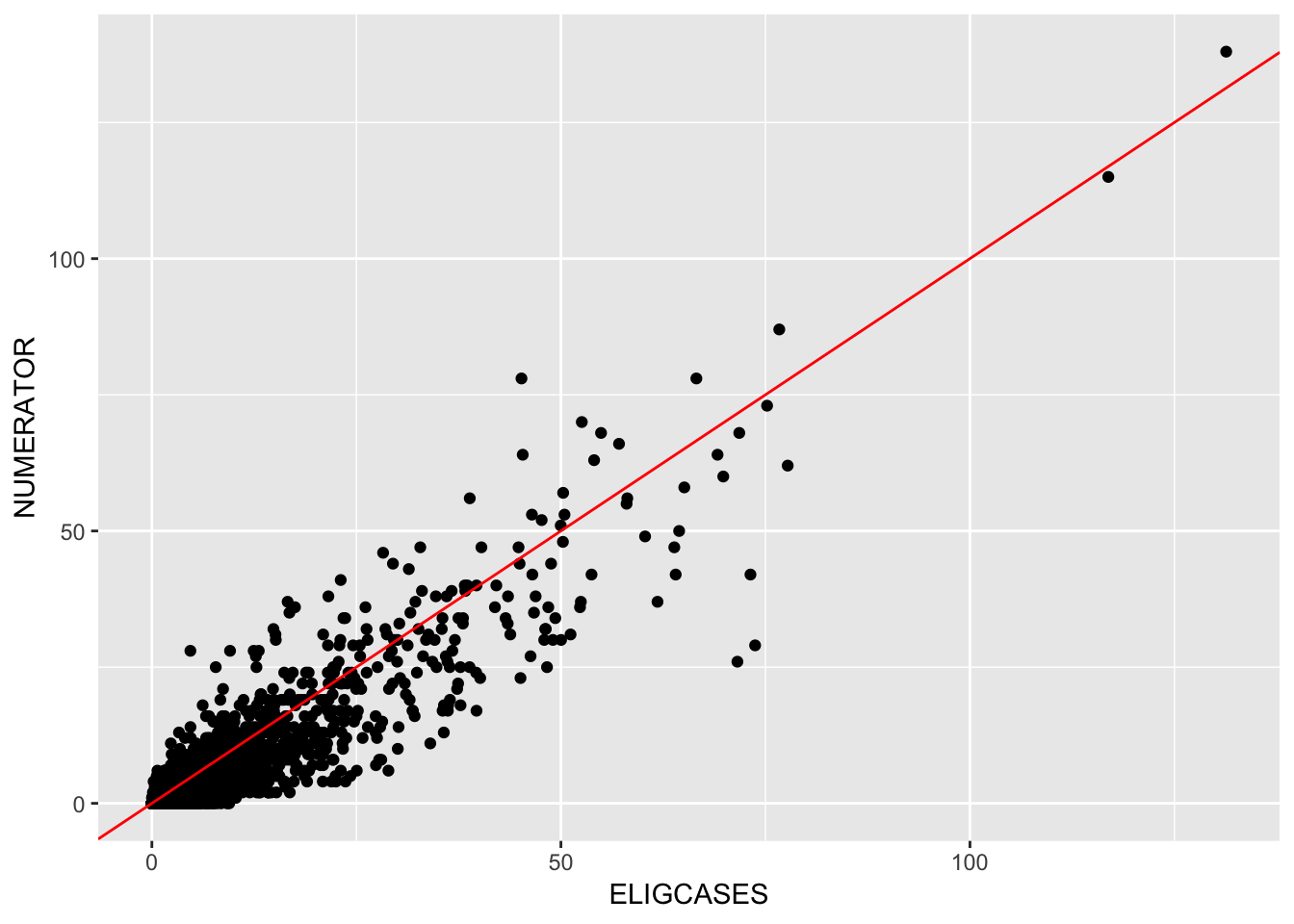
gf_errorbar(CILOWER + CIUPPER ~ sqrt(ELIGCASES), data = Central_Line) ## Warning: Removed 3087 rows containing missing values (geom_errorbar).
CASE STUDY: Which providers to check out …
- Is
ELIGCASEStoo permissive? - Which providers have too high an infection rate?
- Which providers have an admirably low infection rate?
Too_high <-
Central_Line %>%
filter(CILOWER > 1) %>%
select(CILOWER, `Provider ID`) %>%
arrange(desc(CILOWER))
Baddies <-
Institutions %>%
inner_join(Too_high)## Joining, by = "Provider ID"Nice <-
Central_Line %>%
filter(CIUPPER < 1) %>%
select(CIUPPER, `Provider ID`) %>%
arrange(desc(CIUPPER))
Goodies <-
Institutions %>%
inner_join(Nice)## Joining, by = "Provider ID"Activity about confidence intervals: Which under-performing hospitals should we prioritize for inspection? Use the confidence intervals being above 1 to identify them.